 |
| Ribonucleic acid (RNA) |
Ribonucleic acid (RNA), a molecule that plays many roles in the effective usage of genetic information, exists in several forms, each with its own unique function. RNA functions in the process of protein synthesis, during which information from DNA is used to direct the construction of a protein, and possesses enzymatic and regulatory capabilities.
Ribonucleic acid (RNA) is a complex biological molecule that is classified along with deoxyribonucleic acid (DNA) as a nucleic acid. Chemically, RNA is a polymer (long chain) consisting of subunits called ribonucleotides linked together by phosphodiester bonds.
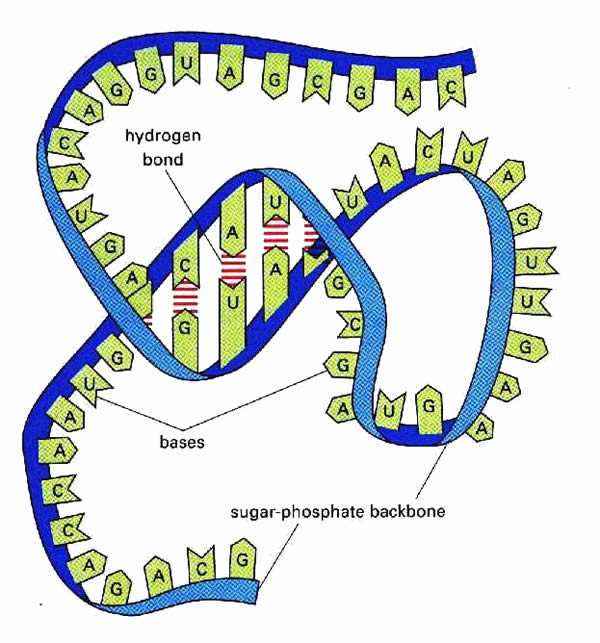
Each ribonucleotide consists of three parts: the sugar ribose (a five-carbon simple sugar), a negatively charged phosphate group, and a nitrogen-containing base. There are four types of ribonucleotides, and the differences between them lie solely in which of four possible bases they contain. The four bases are adenine (A), guanine (G), cytosine (C), and uracil (U).
  |
The structures of DNA and RNA are very similar, but there are three important differences. The sugar found in the nucleotide subunits of DNA is deoxyribose, which is related to but differs slightly from the ribose found in the ribonucleotides of RNA.
In addition, while DNA nucleotides also contain four possible bases, there is no uracil in DNA; instead, DNA nucleotides may contain a different base, called thymine (T). Finally, while DNA exists as a double-stranded helix in nature, RNA is almost always single-stranded.
Folding of RNA Molecules
The significance of many types of RNA lies in the order of their nucleotides, which represents information transcribed from DNA. This nucleotide order is called the primary structure of the molecule. An important aspect of many biological molecules, however, is the way their primary structures fold to create a three-dimensional shape.
A single strand of DNA, for example, associates with another strand in a particular way to form the famous double helix,which represents its actual three dimensional shape in nature. Similarly, protein molecules, especially enzymes, must be folded into a very specific three-dimensional shape if they are to perform their functions; loss of this shape will cause their inactivation.
Since RNA is single-stranded, it was recognized shortly after the discovery of some of its major roles that its capacity for folding is great and that this folding might play an important part in the functioning of the molecule.
The nucleotides in an RNA molecule can form hydrogen-bonded base pairs, according to the same rules that govern DNA base pairing. Cytosine binds to guanine, and uracil binds to adenine. What this means is that in a particular single-stranded RNA molecule, complementary portions of the molecule are able to fold back and form base pairs with one another.
These are often local interactions, and a common structural element that is formed is called a "hairpin loop" or "stem loop." A hairpin loop is formed when two complementary regions are separated by a short stretch of bases so that when they fold back and pair, some bases are left unpaired, forming the loop.
The net sum of these local interactions is referred to as the RNA’s secondary structure and is usually important to an understanding of how the RNA works. All transfer RNAs (tRNAs), for example, are folded into a secondary structure that contains three stem loops and a fourth stem arranged onto a "clover leaf" shape.
Finally, local structural elements may interact with other elements in long-range interactions, causing more complicated folding of the molecule in space. The clover leaf arrangement of a tRNA undergoes further folding so that the entire molecule takes on a roughly L-shaped appearance. An understanding of the three-dimensional shape of an RNA molecule is crucial to understanding its function.
By the late 1990’s, the three-dimensional structures of many tRNAs had been worked out, but it had proven difficult to do X-ray diffraction analyses on most other RNAs because of technical problems. More advanced computer programs and alternate structure-determining techniques have now enabled research in this field to proceed.
Three Classes of RNA
While all RNAs are produced by transcription, several classes of RNA are created, and each has a particular function. By the late 1960’s, three major classes of RNAs had been identified, and their respective roles in the process of protein synthesis had been elucidated.
In general, protein synthesis refers to the assembly of a protein using information encoded in DNA, with RNA acting as an intermediary to carry information and assist in protein building. In 1956, Francis Crick, one of the scientists who had discovered the double-helical structure of DNA, referred to this information flow as the "central dogma," a term that continues to be used.
Messenger RNA (mRNA) is themolecule that carries a copy of the DNA instructions for building a particular protein. It usually represents the information provided by a single gene and carries this information to the ribosome, the site of protein synthesis. This information must be decoded so that it will specify the order of amino acids in a protein.
Nucleotides are read in groups of three (codons). In addition to the information required to order amino acids, the mRNA contains signals that tell the protein-building machinery where to start and stop reading the genetic information.
Ribosomal RNA (rRNA) exists in three distinct sizes and is part of the structure of the ribosome. The three ribosomal RNAs interact with many proteins to complete the ribosome, the cell structure that directs the events of protein synthesis.
One of the functions of the rRNA is to interact with mRNA at a particular location and orient it properly so that reading of its genetic code can begin at the correct location. Another rRNA acts to facilitate the transfer of the growing polypeptide chain from one tRNA to another (peptidyl transferase activity).
Transfer RNA (tRNA) serves the vital role of decoding the genetic information. There are at least twenty and usually fifty to sixty different tRNAs in a given cell. On one side, they contain an "anticodon" loop, which can base-pair to the mRNA codon according to its sequence and the base-pairing rules.
On the other side, they contain an amino acid binding site, to which is attached the appropriate amino acid for its anticodon. In this way, tRNA allows the recognition of any particular mRNA codon and matches it with the appropriate amino acid. The process continues until an entire new protein molecule has been constructed.
Split Genes and mRNA Processing
In bacterial genes, there is a colinearity between the segment of a DNA molecule that is transcribed and the resulting mRNA. In other words, the mRNA sequence is complementary to its template and is the same length, as would be expected.
In the late 1970’s, several groups of scientists made a seemingly bizarre discovery regarding mRNAs in eukaryotes (organisms whose cells contain a nucleus, including all living things that are not bacteria or archaebacteria): The sequences of mRNAs isolated from eukaryotes were not collinear with the DNA from which they were transcribed.
The coding regions of the corresponding DNA were interrupted by seemingly random sequences that served no immediately obvious function. These introns, as they came to be known,were apparently transcribed along with the coding regions (exons) but were somehow removed before them RNA was translated.
This completely unexpected observation led to further investigations that revealed that mRNA is extensively processed, or modified, after its transcription in eukaryotes.
After a eukaryotic mRNA is transcribed, it contains all of the intervening sequences and is referred to as immature, or a "pre-mRNA." Before it can become mature and functional, three major processing events must occur: splicing, the addition of a "cap," and the addition of a "tail."
The process of splicing is a complex one that occurs in the nucleus with the aid of the "spliceosome," a large complex of RNAs and proteins that identify intervening sequences and cut themout of the pre-mRNA.
In addition, the spliceosome must rejoin the sequences from which the intron-encoded nucleotides were removed so that a complete, functional mRNA results. The process must be extremely specific, since a mistake that caused the removal of only one extra nucleotide could change the protein product of translation so radically that it might fail to function.
While splicing is occurring, two other vital events are being performed to make the immature mRNA ready for action. A so-called cap, which consists of a modified guanine, is added to one end of the pre-mRNA by an unconventional linkage. The cap appears to function by interacting with the ribosome, helping to orient the mature mRNA so that translation begins at the proper location.
A tail, which consists of many adenines (often two hundred or more), is also attached to the other end of the pre-mRNA. This so-called poly-A tail, which virtually all eukaryotic mRNAs contain, seems to be involved in determining the relative stability of an mRNA.
These important steps must be performed after transcription in eukaryotes to enable the creation of a mature, functional messenger RNA molecule that is now ready to be translated. Most mRNAs must be transported out of the nucleus before they are used to make proteins, but about 10 to 15 percent of them RNAs produced remain in the nucleus, where they are used to make proteins.
Other Specialized Functions
The traditional roles of RNA in protein synthesis were originally considered the only roles RNA was capable of performing. RNA in general, while considered an important molecule, was thought of as a "helper" in translation.
This all began to change in 1982, when the molecular biologists Thomas Cech and Sidney Altman, working independently and with different systems, reported the existence of RNA molecules that had catalytic or enzyme like activity, meaning that RNA molecules can function as enzymes.
Until this time, it was believed that all enzymes were protein molecules. The importance of these findings cannot be overstated, and Cech and Altman ultimately shared the 1989 Nobel Prize in Chemistry for the discovery of these RNA enzymes, or ribozymes.
Both of these initial ribozymes catalyzed reactions that involved the cleavage of other RNA molecules—that is, they acted as nucleases. Subsequently, many ribozymes have been found in various organisms, from bacteria to humans. Some of them are able to catalyze different types of reactions, and there are new ones reported every year.
Thus ribozymes are not a mere curiosity but play an integral role in the molecular machinery of many organisms. Their discovery also gave rise to the idea that at one point in evolutionary history, molecular systems composed solely of RNA performing many roles existed in an "RNA world."
At around the same time as these momentous discoveries, still other classes of RNAs were being discovered, each with its own specialized functions. In 1981, Jun-Ichi Tomizawa discovered RNAI, the first example of what would become another major class of RNAs, the antisense RNAs.
The RNAs in this group are complementary to a target molecule (usually an mRNA) and exert their function by binding to that target via complementary base pairing. These anti sense RNAs usually play a regulatory role, often acting to prevent translation of the relevant mRNA to modulate the expression of the protein for which it codes.
Another major class of RNAs, the small nuclear RNAs (snRNAs), was also discovered in the early 1980’s. Molecular biologist Joan Steitz was working on the autoimmune disease systemic lupus when she began to characterize the snRNAs.
There are six different snRNAs, now called U1-U6 RNAs. These RNAs exist in the nuclei of eukaryotic cells and play a vital role in mRNA splicing.
They associate with proteins in the spliceosome, forming so-called ribonucleoprotein complexes (snRNPs), and play a prominent role in detecting proper splice sites and directing the protein enzymes to cut and paste at the proper locations.
It has been known since the late 1950’s that many viruses containRNA, and not DNA, as their genetic material. This is another fascinating role for RNA in the world of biology.
The viruses that cause influenza, polio, and a host of other diseases are RNA viruses. Of particular note are a class of RNA viruses known as retroviruses because they have a particularly interesting life cycle.
Retroviruses, which include human immunodeficiency human immuno deficiency virus (HIV), the virus that causes acquired immuno deficiency syndrome (AIDS) in humans, use a special enzyme called reverse transcriptase to make a DNA copy of their RNA instructions when they enter a cell.
That DNA copy is inserted into the DNA of the host cell, where it is referred to as a provirus, and never leaves. Clearly, understanding the structures and functions of the RNAs associated with these viruses will be important in attempting to create effective treatments for the diseases associated with them.
An additional role of RNA was noted during the elucidation of the mechanism of DNA replication. It was found that a small piece of RNA, called a primer, must be laid down by the enzyme primase before DNA polymerase adds DNA nucleotides to this initial RNA sequence, which is subsequently removed.