 |
| Ribosome |
Ribosomes are complex cellular structures found in all cells and are responsible for making proteins. They are composed of ribosomal RNA (rRNA) and protein and are most abundant in the cytoplasm, although some functional ribosomes can be found in the nuclei of eukaryotic cells. Chloroplasts and mitochondria also have their own ribosomes.
Ribosomes are responsible for synthesizing the proteins in all cells by a process called translation. It is called translation because ribosomes use messenger ribonucleic acids (mRNAs) as their guide and must "translate" the message contained in the nucleotides of mRNAs.
The general structure of ribosomes is the same in all cells, but ribosomes of prokaryotes are smaller than ribosomes in the cytoplasm of eukaryotes. The ribosomes in chloroplasts and mitochondria are more similar to the smaller ribosomes of prokaryotes but are often smaller yet.
  |
Ribosome Structure
Because ribosomes, and rRNAs, are so large, they are not described on the basis of their molecular weight but rather in Svedbergs, or Svedberg units.
Svedbergs (denoted S) are a measure of how quickly a large molecule or aggregation of molecules sediment, or sink to the bottom of a centrifuge tube while being spun around. Higher S values mean faster sedimentation and thus greater mass. For example, prokaryotic ribosomes sediment at 70’s, whereas eukaryotic ribosomes, which are larger, sediment at 80’s.
Svedbergs cannot be added together, because they are not directly related to mass but to a combination of mass and overall molecular size and shape. Consequently, if two molecules that are both 30’s are joined together, their combined sedimentation rate would be about 50’s or a little more but not 60’s.
Prokaryotic ribosomes constitute three rRNAs and fifty-two different proteins. Like all ribosomes, they are composed of two subunits, referred to simply as large and small subunits.
Large subunits have a sedimentation rate of 50’s and are composed of a 23S rRNA, a 5S rRNA, and thirty-one proteins. Small subunits have a sedimentation rate of 30’s and are composed of a single 16S rRNA and twenty-one different proteins. To function, a small and large subunit must come together.
Eukaryotic ribosomes are more complex, with four rRNAs and more than eighty proteins. Large subunits have a sedimentation rate of 60’s and are composed of a 28S rRNA, a 5.8S rRNA, a 5S rRNA, and about forty-nine proteins. Small subunits have a sedimentation rate of 40’s and are composed of a single 18S rRNA and about thirty-three proteins.
Assembly of the subunits takes place in a region of the nucleus called the nucleolus. When completed, the subunits are transported through nuclear pores to the cytoplasm. Although translation of mRNAs takes place primarily in the cytoplasm, a small amount occurs in the nucleus.
Translation in Prokaryotes
Before translation can take place, transcription of a gene must occur. Transcription converts the deoxyribonucleic acid (DNA) code of a gene into a complimentary RNA code, in the form of an mRNA. Even while RNA polymerase catalyzes the joining of nucleotides for the mRNA, translation can begin.
In initiation, special proteins called initiation factors (IFs) enable the small subunit of a ribosome to bind to an mRNA and form the initiation complex. Next, a special tRNA called fMet-tRNA(N-formylmethionyl-tRNA), which carries a specially modified amino acid derivative of methionine, binds to the start codon of the mRNA(AUG).
This modified methionine is the first amino acid in all prokaryotic proteins, but many proteins are modified after transcription, often by removing the first few amino acids. Lastly, the large ribosomal subunit binds.
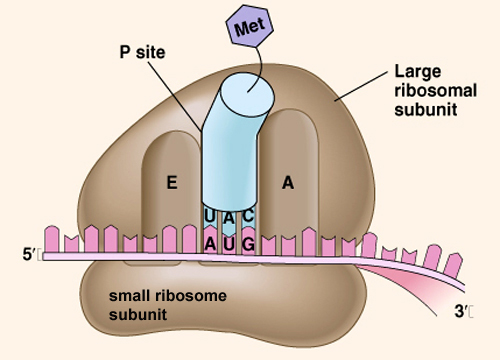
Elongation is a complex process in which the ribosome adds amino acids, one at a time, to the growing protein. Ribosomes have three sites where events in translation takes place. The A site (aminoacyl-tRNA site) is where new tRNAs, with their attached amino acids, bind to the ribosome and mRNA.
The P site (peptidyl-tRNAsite) is where the tRNA with the growing protein (polypeptide) is attached to the ribosome and them RNA. The E site (exit site), only recently recognized, is where tRNAs leave the ribosome after they have completed their work.
The sequence of events in elongation can be difficult to visualize, but it is a cyclical process that repeats until elongation is done. The steps are as follows.
First, while the tRNA with the growing protein (called a peptidyl-tRNA) rests in the Psite, a tRNA with an amino acid (called an aminoacyl-tRNA) enters the A site and binds to the codon on the mRNA. Second, the peptidyl-tRNA attaches the growing protein to the amino acid on the aminoacyl-tRNA. Now the tRNA in the P site has no amino acids attached to it.
Third, the tRNA in the P site now leaves the ribosome by passing through the E site. Fourth, as the tRNA leaves the P site, the tRNA at the A site (now a peptidyl-tRNA) is translocated (moved) to the P site by moving the mRNA over one codon, leaving the A site open for the next aminoacyl-tRNA to attach.
These steps repeat until the stop codon, near the end of the mRNA, is reached. Many of the events of elongation were long believed to be catalyzed by proteins in the ribosome. It is now known that some of the ribosome’s catalytic properties are due to the rRNAs, which, as a result, are sometimes called ribozymes.
The stop codon is not recognized by any of the tRNAs. Termination occurs when some special proteins called releasing factors bind to the ribosome. When they bind, the protein that is attached to the tRNA in the P site is released from the tRNA, the tRNA leaves through the E site, and the two subunits come apart.
Translation in Eukaryotes
The process of translation in eukaryotes varies only in minor details from translation in prokaryotes, and these differences are due to the greater complexity of eukaryotic mRNA and ribosomes.
Eukaryotic mRNA must go through extensive processing after being transcribed, including intron excision/exon splicing and addition of a special 5′ cap Translation includes three steps: initiation, elongation, and termination.
and a 3′ poly-adenosine tail. Eukaryotic mRNA cannot be translated until these modifications are completed. Although some mRNA is translated in the nucleus, most must be transported to the cytoplasm first. Consequently, transcription and translation are completely separate processes, unlike their coupling in prokaryotes.
Initiation in eukaryotes involves a larger number of initiation factors. Once the small ribosomal subunit binds, it then starts at the 5′ end of the mRNA and searches for the start codon (AUG). When the start codon is found, a methionyl-tRNA binds to the ribosome and the start codon of the mRNA.
In eukaryotes, like in prokaryotes, the first amino acid in all proteins is methionine, but in eukaryotes it is not a N-formyl-methionine. Elongation and termination are essentially the same in both eukaryotes and prokaryotes. As in prokaryotes, completed proteins are typically modified in various ways before being used by the cell.
Many ribosomes will typically translate an mRNA simultaneously. An mRNA with several ribosomes lined up along it, all of them translating at once, is often called a polyribosome. Polyribosomes are observed in both eukaryotes and prokaryotes.
Distribution in Eukaryotic Cells
After being synthesized in the nucleolus, a few ribosomes will function for an unknown length of time in the nucleus. Most, though, are transported to the cytoplasm as separate subunits. Once in the cytoplasm, ribosomes will either bind to endoplasmic reticulum (ER), in regions called rough ER because of the fuzzy appearance of these areas in electron micrographs, or they will remain free in the cytoplasm.
Ribosomes bound to the rough ER specialize in making proteins that will either be embedded inmembranes or transported in vesicles to the Golgi complex. The different fates of proteins made at the rough ER are determined by special signal sequences, sequences of amino acids at the beginning of proteins which are typically removed later.
Ribosomes that are free in the cytoplasm make proteins of various kinds that are needed for processes occurring in the cytoplasm. Regardless of location, ribosomes are usually found disassembled, and the two subunits come together only when needed for translation. Cells have anywhere from several thousand ribosomes to as many a few million in cells that have particularly high rates of protein synthesis.